SPECIAL SESSIONS
In addition to the main track, the conference will provide for a number of special sessions focused on specific, cutting-edge topics. Click on the name of the special session to expand.
Machine-learning and statistical techniques are at the base of many classifiers or predictive
models, often employed inside Decision Support Systems. Among them, Clinical Decision Support
Systems (CDSSs), used to support decision-making activities in healthcare, operate in critical
scenarios where it is fundamental to have full knowledge, reliability and explainability of the IT
process.
In human health management, it is no longer sufficient to use ICT tools as ‘black-boxes,’ without
providing insights about how data are processed and quantified. For this reason, the explainability
of the whole processing chain is a desiderable and fundamental feature in clinical environments.
What happens in each step, which entities are involved, and what their role in the
model’s/classifier’s outcome is are mandatory requirements. The benefits of implementing explainable
models are many: user acceptance, control, trustworthiness, insightfulness, ethical and legal
aspects, explanatory debugging.
The explainability of a predictive model or classifier must be planned in the design phase, because
not all methodologies are natively explainable. Some focal points that must be taken into account,
focusing on the used algorithm and the input data. To make a model explainable, a multi-level
strategy can be employed considering all the stakeholders involved in the process: the developer
needs to verify the functionality learned from the model; the clinicians need the necessary tools to
confirm experimental evidence; and the patient, whose health is entrusted to the response of the
system, must be informed with an explanation about the results. In this special session, research
papers proposing predictive models in clinical activity, dealing with feature interpretability and
model explainability, are welcome.
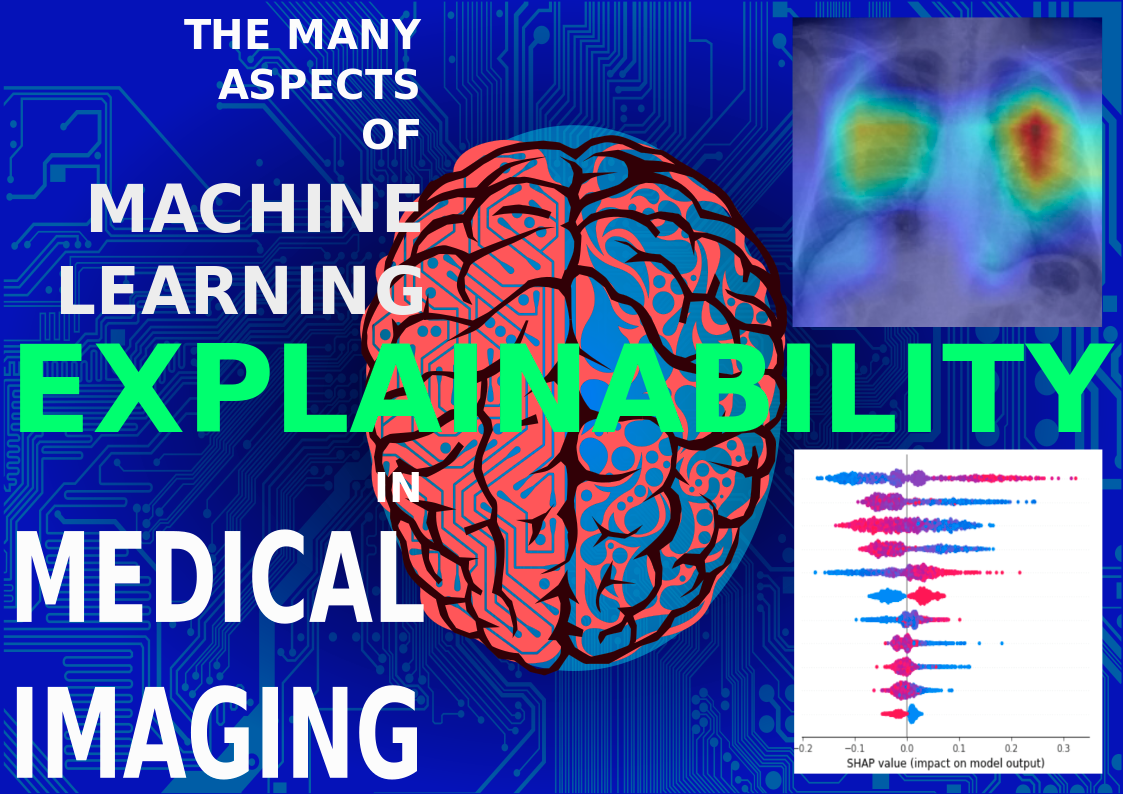
Research areas include, but are not limited to:
- - Explainable machine learning
- - Visual analytics for explainable machine learning
- - Interpretable features
- - Decision-making support in healthcare
- - Explainable predictive models
Special session organisers:
- Salvatore Calderaro , Department of Mathematics and Informatics (DMI), University of Palermo, Italy
- Carmelo Militello , Institute for High-Performance Computing and Networking, National Research Council (ICAR-CNR), Palermo, Italy
- Francesco Prinzi , Department of Biomedicine, Neuroscience and Advanced Diagnostics (BiND), University of Palermo, Palermo, Italy
- Filippo Vella , Institute for High-Performance Computing and Networking, National Research Council (ICAR-CNR), Palermo, Italy
Machine learning has become a pivotal tool to analyse biomedical and biological datasets, especially
in the Big Data era. In fact, machine learning algorithms can identify hidden relationships and
structures in health care data, and even take advantage of them to formulate accurate predictions
about similar or future data instances. For example, machine learning software has been able to
predict the diagnosis of oncological patients just by processing patients' clinical features,
allowing scientists to save time and money compared to wet lab experiments. Computational
researchers have also exploited machine learning to infer knowledge about patients by analysing
biological datasets, especially the ones featuring genetics and epigenomic traits. Data mining
approaches applied to such datasets, in fact, can lead to relevant discoveries both to understand
molecular biology and to gain new knowledge about patients’ diseases.
Our special session on "Machine Learning in Healthcare Informatics and Medical Biology" aims at
boosting these scientific fields, calling for researchers able to show the potential and the
advances of machine learning algorithms, in making accurate computational predictions in health care
datasets and in patient-oriented biological datasets.

Topics of interest include:
- - Machine learning methods applied to health care and biomedical datasets
- - Machine learning methods applied to genetics and epigenomics datasets, to understand the conditions of healthy and/or sick patients
- - Machine learning methods applied to biological datasets to understand the underlying biomolecular scenario
- - Machine learning software and tools in the health care and biological domains
- - Statistical models to analyse health care, biomedical, and biological datasets
- - Data mining applications in the health care and biological domains
Special session organisers:
- Davide Chicco , University of Toronto, Canada
- Giuseppe Jurman , Fondazione Bruno Kessler, Italy
- Giuseppe Agapito , Università Magna Graecia di Catanzaro, Italy
- Joao Ribeiro Pinto , Bosch, Portugal
- Abbas Alameer , Kuwait University, Kuwait
We know that cells are not homogenous objects, as they are composed of membranes, cytoplasm, nuclei,
and various organelles. Therefore, their precise single cell discrimination in different classes
and/or states via Machine Learning is highly challenging. To prevent wrong predictions of a Machine
Learning model, we should better understand the confidence of each individual prediction.
For instance, Machine Learning is well suited to find patterns in large cell sequencing datasets.
But how can we quantify uncertainty in genomics applications? Furthermore, what happens if a model
must predict an out-of-distribution cell in an open-world scenario? In other words, a prediction
model should first define a certain in-distribution (test cell is assumed to be drawn from the
training data of known cells), to detect a certain out-of-distribution sample. In fact, a model can
cause unsafe prediction failures due to its blindness and poor awareness. Moreover, it conveys low
confidence in its correctness, which can be problematic for a wide range of Biomedical and Health
informatics applications.
For instance, standard metrics for the evaluation of Machine Learning models measure overall model
performance on a whole cell dataset and provide no indication of the model confidence in the
correctness of an individual prediction on a new cell. Furthermore, model performance metrics cannot
be easily updated after initial model testing. The mentioned limitations can result in prediction
failure, when a model is faced with out-of-distribution data. Hence, how can we detect unknown
cells? How precise are genomic data patterns?
The scope of this special session is to invite the research community to reason about the following
open questions: how they measure out-of-distribution events; how we can better predict unknown
cells; which parameters can help the Biomedical Community to clearly identify out-of-distribution
events; how we can quantify the uncertainty of Machine Learning patterns in cell sequencing
applications; and how this could influence the future of precision medicine.

Topics of interest include, but are not limited to:
- - Out-of-distribution recognition for single cell applications
- - Uncertainty in cell sequencing
- - Metrics for model evaluation and prediction
- - Unknown single cell/sample prediction techniques
- - Mathematical modelling for out-of-distribution recognition
- - Open-set versus closed-set recognition
- - Prediction uncertainty in the Biomedical field
- - Methods to visualise out-of-distribution events
- - Application of out-of-distribution recognition in the Biomedical field
Special session organisers:
In the last decade, many efforts have been devoted to developing new technologies able to improve
the understanding of the molecular mechanisms of cells. These cutting-edge technologies are often
used in a broad cellular context and give rise to complex data that are difficult to interpret and
correlate.
Population-based sequencing approaches (aka bulk RNA-seq, ChIPseq, ATACseq) have played a
significant role in decoding genome-wide transcriptomics and epigenomics variations across a broad
range of fields, including oncology, stem cell biology, immunology, and neuroscience. However, as
bulk data represents an average of feature expression across a population of cells, it may hide the
transcriptional trends of distinct subpopulations with the most abundant cell types or states.
To increase specificity and efficiency, multiple technologies for single-cell inspection arose, such
as transcriptomics (i.e. scRNAseq) and epigenomics (i.e. scATACseq), allowing the study of cellular
mechanisms. Finally, nowadays a huge interest is evolving around the investigation of in-vivo
derived tissues thanks to the birth of single-cell spatial transcriptomics and single-cell
multimodal technologies, providing even finer levels of resolution. These single-cell sequencing
technologies offer unprecedented opportunities for exploring new computational, mathematical, and
statistical approaches capable of solving problems related to the discovery of novel biological
insights.
The scope of this special session is to bring together researchers involved in the development of
methods and approaches for the analysis and integration of single-cell data, with a particular focus
on recently born advanced technologies.

Topics of interest include:
- - Artificial intelligence and machine learning for the discovery of novel cell types and proteome profiling
- - Trajectories methods for studying cellular dynamic processes at a previously unattainable spatial and temporal resolution
- - Network-based methodologies featuring in-cell variation such as gene and cell interactions
- - Clustering techniques for single-cell omics data
- - Integration approaches for multimodal data
- - Analysis methods for single-cell spatial transcriptomics
- - Novel tools for analysis and integration of single-cell omics data
Invited speaker: Davide Risso
Special session organisers:
- Annamaria Carissimo , Institute for Applied Mathematics of the National Council for Research (IAC-CNR), Napoli, Italy
- Dario Righelli , University of Padova, Padova, Italy
This special session aims at providing a picture of the current research and future perspective,
which Italian research institutes of Informatics, and their partners, are currently performing in
bioinformatics.
The national CINI (Consorzio Interuniversitario Nazionale di Informatica) InfoLife laboratory
promotes networking initiatives among researchers with an informatics background who conduct
relevant research in bioinformatics, and related topics, together with their international partners.
Italy is an important source and partner in providing computational approaches for any aspect of the
bioinformatic field, from the development of baseline specialised algorithms and data structures to
the implementation of high-level data analysis and visualisation.
This special session aims at being an opportunity for Italian researchers and their partners to
expose their current and future research directions, and for other scientists to meet such a
community.

Relevant topics include:
- - Computational transcriptomics and spatial transcriptomic data analysis
- - Single-cell omics
- - Multi-omics data analysis and models
- - Computational pangenomics
- - High-performance computing for omics data
- - Algorithms and data structures for bioinformatics
- - Knowledge management in bioinformatics and medical informatics
- - Drug development and pharmacovigilance
- - Artificial intelligence and machine learning in bioinformatics
- - Advanced technique for bio-medical content visualisation
- - Bio-inspired computing for bioinformatics
- - Computational modelling and omics data
Invited speaker: Marco Beccuti
Special session organisers:
- Vincenzo Bonnici , University of Parma, Parma, Italy
- Giacomo Baruzzo , University of Padova, Padova, Italy
- Simone Pernice , University of Torino, Torino, Italy
Distributed computing systems have become pivotal in the analysis of bioinformatics data, especially
for the investigation of complex biological problems that require intense computational workloads.
The usage of distributed computing systems like Apache Spark, in fact, might allow scientific
discoveries on bioinformatics data that otherwise would be impossible if the analyses were performed
on personal computers.
Our special session on "Distributed computing in bioinformatics and computational biology" aims at
calling for researchers able to show the potential and the advance of distributed computing
resources to make accurate computational analyses in biological datasets.
With this special session, we would like to gather scientific contributions aimed at optimising the
usage of distributed computing systems and resources in any bioinformatics, genomics, and proteomics
context. The results presented in these studies would be useful and helpful both for the distributed
computing experts and for inexperienced users approaching this subject for the first time.
Topics of interest include:
- - Distributed computing systems in genomics and proteomics
- - Bioinformatics projects enhanced by distributed computing platforms
- - Optimisation of distributed computing systems
- - Application of machine learning methods to bioinformatics datasets enhanced through distributed computing
- - Protocols for efficient usage of distributed computing systems
Special session organisers:
- Davide Chicco , University of Toronto, Canada
- Umberto Ferraro Petrillo , Sapienza Università di Roma, Italy
- Giuseppe Cattaneo , Università di Salerno, Italy
- Raffaele Giancarlo , Università di Palermo, Italy
- Lorenzo Di Rocco , Sapienza Università di Roma, Italy
- Giorgio Grani , Sapienza Università di Roma, Italy
Computational biology is a field that involves the analysis of biological systems at various levels
of complexity, using appropriate modelling frameworks and computational methods.
With the advancement of computational biology approaches and modelling systems, the current
challenge is to employ these techniques to define personalised models that identify tailored drugs
and therapies, thus actualising the personalised medicine paradigm. The aim of this special session
is to bring together researchers who are working on the development of methods and models applied in
the fields of computational biology and systems medicine to facilitate the exchange of knowledge and
ideas in these innovative and fundamental research areas.
Topics of interest include:
- - Parameterisation and verification of biomedical models
- - Modelling neural activity
- - Individual-aware models to assess the impact of genetic variation on cellular regulatory network
- - Cancer progression models
- - Epidemiological models
- - Multiscale models and simulation of biological systems
- - Space-temporal models and simulation of biological systems
- - Robustness of cellular networks
- - Emergent properties in complex biological systems
- - Metabolic and signalling pathways analysis and engineering
- - Genetic variants impact on epigenetic elements
- - Models for personalised and targeted therapies
- - Patients classification and stratification
- - Drug combination, repositioning, and recommendation for personalised medicine
- - Clinical data integration into systems biology models.
Invited speaker: Marco Antoniotti
Special session organisers:
- Chiara Damiani , University of Milano-Bicocca, Italy
- Bruno Giovanni Galuzzi , University of Milano-Bicocca, Italy
- Vincenzo Bonnici , University of Parma, Parma, Italy
- Marco Beccuti , University of Turin, Italy
- Rosalba Giugno , University of Verona, Italy
- Manuel Tognon , University of Verona, Italy
- Simone Avesani , University of Verona, Italy
- Eva Viesi , University of Verona, Italy